Exceptional performance in the general population
The Harmony test outperformed traditional first-trimester screening (FTS) in a landmark study (NEXT) published in the New England Journal of Medicine.2 In this prospective blinded study, the Harmony test was found to be superior to FTS in terms of detection rate, false positive rate and positive predictive value for trisomy 21.
Watch video by Ron Wapner, MD, co-author of the NEXT study
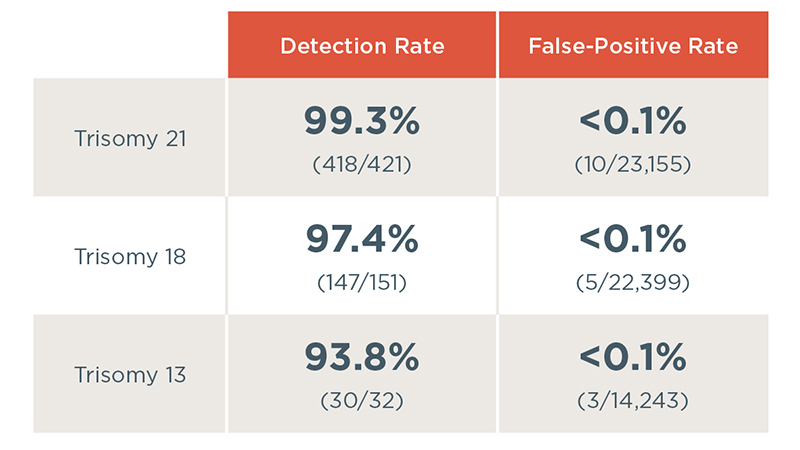
Performance of the Harmony test
The Harmony test delivers consistent, industry-leading performance for trisomy 21, trisomy 18 and trisomy 13 across clinical studies.3
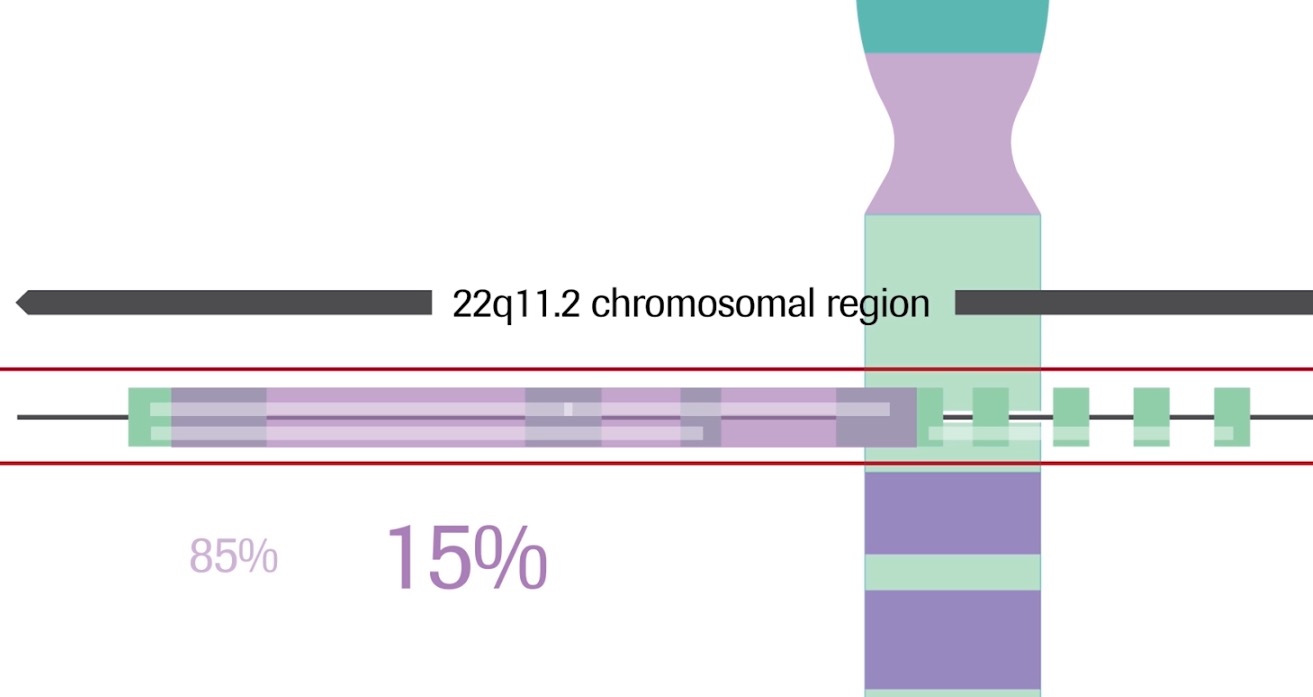
22q11.2 microdeletion
22q11.2 microdeletion, an absence of a small piece of chromosome 22,4 happens in about 1 in 1000 pregnancies.5,6 It is the second most common genetic cause of heart defects and developmental delay after Down syndrome,7 and is the underlying cause of DiGeorge and velocardiofacial syndromes (VCFS). 22q11.2 microdeletion is not reliably detected by routine prenatal screening or karyotype.7
Unlike trisomies, maternal age is not a risk factor for the microdeletion5 and family history cannot reliably predict its occurrence, as more than 90% of affected individuals have no family history.8
The performance of the Harmony test for 22q11.2 microdeletion has been evaluated in a study including over 1900 samples, including 129 with confirmed deletions. The Harmony test was able to reliably identify pregnancies at risk for 22q11.2 deletions of 3Mb and smaller with a low false positive rate.5
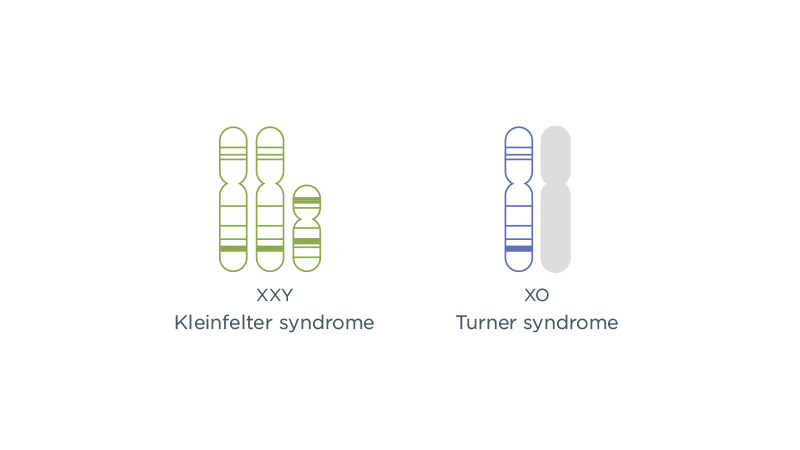
Sex chromosome aneuploidies
Individuals typically have two X chromosomes or one X and one Y chromosome. Any variation from the typical number of X and Y chromosomes in the cells is called a sex chromosome aneuploidy (SCA). SCAs have a combined prevalence of about 1:500.9 Conditions associated with SCAs have widely variable clinical features that are often subtle but can include birth defects, infertility and learning differences. Most SCAs are not reliably detected by routine prenatal screening.
Twin pregnancies
Performance of the Harmony test has been published in peer-reviewed studies including over 1,300 twin pregnancies and has been used in over 16,000 twin pregnancies across the world.10,11,12,13,14,15,16
Clinical scenarios included in published Harmony twin studies
- Monochorionic twins
- Dichorionic twins
- Twins conceived with in vitro fertilization (IVF)
- Twins conceived naturally
- Pregnancies where both twins have aneuploidy
- Pregnancies where only one twin has aneuploidy
- First trimester testing
- Second trimester testing
Professional societies such as The International Society for Prenatal Diagnosis (ISPD) and American College of Medical Genetics and Genomics (ACMG) support offering NIPT as a first-line screen.
“The following protocol options are currently considered appropriate: cfDNA screening as a primary test offered to all pregnant women.”17
“ACMG recommends informing all pregnant women that NIPS is the most sensitive screening option for trisomy 21, trisomy 18, and trisomy 13.”18
References
- Norton et al. New Engl J of Med. 2015; 372(17):1589-1597.
- Norton et al. Am J Obstet Gynecol.2012 Aug;207(2):137-8.
- Stokowski et al. Prenat Diagn. 2015 Dec;35(12):1243-6.
- Schmid et al. Fetal Diagn Ther 2017; Doi:10.1159/000484317.
- Grati et al. Prenat Diagn 2015;35:801-809.
- Wapner et al. N Engl J Med 012;367:2175-2184.
- McDonald-McGinn et al. Gnet Med.2001 Jan-Feb:3(1):23-9.
- Bassett et al. JPediatr. 2011 Aug;159(2):332-9.
- Thompson and Thompson. Genetics in Medicine, Sixth Edition. Robert L Nussbaum, Roderick Mclnnes, Willard Huntington, Saunders, 2001.
- del Mar Gil M, et al. Fetal Diagn Ther. 2014;35(3):204-211.
- Struble C et al. Fetal Diagn Ther. 2013;35(3):199-203.
- Bevilacqua E, et al. Ultrasound Obstet Gynecol. 2015;45(1):61-66.
- Stokowski et al. Prenat Diagn. 2015 Dec;35(12):1243-6.
- Sarno L, et al. Ultrasound Obstet Gynecol. 2016;47(6):705-711.
- Jones KJ, et al. Obstet Gynecol. 2018 51:274-277.
- Data on file.
- Benn et al. Prenat Diagn 2015; 35: 725–734.
- Gregg et al. Genetics in Medicine 2016 Oct; 18(10):1056-65.